import logging
import numpy as np
import pandas as pd
import weka.core.jvm as jvm
import weka.plot.classifiers as plcls
from weka.classifiers import Classifier, Evaluation
from weka.core.classes import Random
from weka.core.converters import Loader
%matplotlib widget
jvm.start(logging_level=logging.ERROR)Class imbalance problem¶
In this notebook, we will analyze a skewed dataset for detecting microcalcifications in mammograms. The goal is to build a classifier to identify whether a bright spot in a mammogram is a micro-calcification (an early sign of breast cancer).
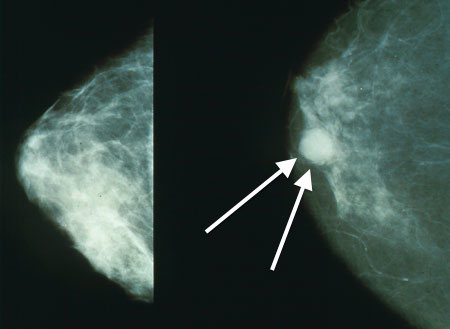
Figure 1:Micro-calcification
The dataset can be downloaded from
OpenML in ARFF format. The following loads the data using python-weka-wrapper.
loader = Loader(classname="weka.core.converters.ArffLoader")
data = loader.load_url("https://www.openml.org/data/download/52214/phpn1jVwe")
data.class_is_last()
print(data.summary(data))There are 7 attributes and over 11 thousand instances. To understand the dataset, refer to Section 4 of the original paper (Woods et al. 1993):
To compute the 10-fold cross-validation accuracy for J48:
clf = Classifier(classname="weka.classifiers.trees.J48")
evl = Evaluation(data)
evl.crossvalidate_model(clf, data, 10, Random(1))
print(f"Accuracy: {evl.percent_correct:.3g}%")You should see that the accuracy is close to 100%. To show the confusion matrix:
confusion_matrix = pd.DataFrame(
evl.confusion_matrix,
dtype=int,
columns=[f'predicted class "{v}"' for v in data.class_attribute.values],
index=[f'class "{v}"' for v in data.class_attribute.values],
)
confusion_matrixEach row of the confusion matrix corresponds to a class value (1: malignant, -1: benign), and each column corresponds to a predicted class. Each entry is a count of instances belonging to a specific class and having a particular predicted class.
# YOUR CODE HERE
raise NotImplementedError()
print(f"Percentage of malignant detected: {percent_of_malignant_detected:.3g}%")# testsDifferent Performance Metrics¶
For a skewed dataset, one can achieve very high accuracy even by ZeroR, i.e., also predicting the class as the majority class regardless of the values of the input features.
We must use other performance metrics to train and evaluate a classification algorithm properly.
To show the above metrics:
pos_class = 1 # specify the postive class value
performance = {
"precision": evl.precision(pos_class),
"recall": evl.recall(pos_class),
"specificity": evl.true_negative_rate(pos_class),
}
performanceAlthough specificity is close to 100%, precision and recall are below 80% and 60% respectively:
- If a bright spot is classified as malignant, the chance it is malignant is less than 80%.
- Out of all malignant bright spots, less than 60% are identified as malignant.
The reason why close to 100% benign bright spots are identified as benign
- is mainly because most bright spots are benign, but
- not because the classifier can distinguish malignant bright spots from benign ones.
TP = evl.num_true_positives(pos_class)
FN = evl.num_false_negatives(pos_class)
FP = evl.num_false_positives(pos_class)
TN = evl.num_true_negatives(pos_class)
assert np.isclose(performance["precision"], TP / (TP + FP))
assert np.isclose(performance["recall"], TP / (TP + FN))
assert np.isclose(performance["specificity"], TN / (TN + FP))
TFPN = pd.DataFrame(
[[TP, FN], [FP, TP]],
dtype=int,
columns=["predicted +ve", "predicted -ve"],
index=["+ve", "-ve"],
)
TFPNThe above table is not the same as a confusion matrix since a confusion matrix
- does not specify a positive class, and
- can have more than two rows/columns in multi-class classification problems.
# YOUR CODE HERE
raise NotImplementedError()
print(f"negative predictive value (NPV): {performance['NPV']:.3g}")# tests-score is another measure that captures the performance in both precision and recall:
-score is useful in training a classifier to maximize both precision and recall.
performance["F"] = evl.f_measure(pos_class)
print(f"F-score: {performance['F']:.3g}")# YOUR CODE HERE
raise NotImplementedError()
print(f"F_2 score: {performance['F_2']:.3g}")# YOUR CODE HERE
raise NotImplementedError()
ZeroR_performanceYOUR ANSWER HERE
Operating Curves for Probabilistic Classifier¶
For a probabilistic classifier that returns probabilities of different classes, we can obtain a trade-off between precision and recall by changing a threshold γ for positive prediction, i.e., predict positive if and only if the probability estimate for positive class is larger than γ.
To plot the precision-recall curve and prints the area under the curve, we can use the following tool:
import weka.plot.classifiers as plclsplcls.plot_prc(evl, class_index=[1])
performance["PRC"] = evl.area_under_prc(pos_class)
print(f"area under precision-recall curve (PRC): {performance['PRC']:.3g}")YOUR ANSWER HERE
YOUR ANSWER HERE
We can also plot the ROC (receiver operator characteristics) curve to show the trade-off between recall (true positive rate) and false positive rate:
plcls.plot_roc(evl, class_index=[1])
performance["AUC"] = evl.area_under_roc(pos_class)
print(f"area under ROC curve (AUC): {performance['AUC']:.3g}")YOUR ANSWER HERE
- Woods, K. S., Solka, J. L., Priebe, C. E., Kegelmeyer, W. P., Doss, C. C., & Bowyer, K. W. (1994). Comparative Evaluation of Pattern Recognition Techniques for Detection of Microcalcifications in Mammography. In State of the Art in Digital Mammographic Image Analysis (pp. 213–231). WORLD SCIENTIFIC. 10.1142/9789812797834_0011