import ipywidgets as widgets
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import seaborn as sns
from ipywidgets import interact
from sklearn import datasets, preprocessing
from sklearn.cluster import DBSCAN
from sklearn.pipeline import make_pipeline
%matplotlib inlineDBSCAN with scikit-learn¶
DBSCAN (Density-based spatial clustering of applications with noise) is a clustering algorithm that identifies clusters as regions of densely populated instances.
We will create synthetic datasets using the sample generators of sklearn. In particular, we first create spherical clusters using sklearn.datasets.make_blobs:
def XY2df(X, Y):
"""Return a DataFrame for 2D data with 2 input features X and a target Y."""
df = pd.DataFrame(columns=["feature1", "feature2", "target"])
df["target"] = Y
df[["feature1", "feature2"]] = X
return df
fig, ax = plt.subplots(clear=True, figsize=(10, 10), layout="constrained", num=1)
ax.set_aspect("equal")
@interact
def generate_blobs(
n_samples=widgets.IntSlider(value=200, min=10, max=1000, continuous_update=False),
centers=widgets.IntSlider(value=3, min=1, max=4, continuous_update=False),
cluster_std=widgets.FloatSlider(
value=0.5, min=0, max=5, step=0.1, continuous_update=False
),
):
df = XY2df(
*datasets.make_blobs(
n_samples=n_samples,
centers=centers,
cluster_std=cluster_std,
random_state=0,
)
)
ax.clear()
sns.scatterplot(data=df, x="feature1", y="feature2", hue="target", ax=ax)
plt.show()We will use the dataset df_spherical created with the default parameters specified above:
df_spherical = XY2df(
*datasets.make_blobs(n_samples=200, centers=3, cluster_std=0.5, random_state=0)
)To create non-spherical clusters, one way is to use sklearn.datasets.make_circle.
df_nonspherical = XY2df(
*datasets.make_circles(n_samples=200, factor=0.1, noise=0.1, random_state=0)
)fig, ax = plt.subplots(clear=True, figsize=(10, 10), layout="constrained", num=2)
ax.set_aspect("equal")
@interact
def generate_circles(
n_samples=widgets.IntSlider(value=200, min=10, max=1000, continuous_update=False),
factor=widgets.FloatSlider(
value=0.1, min=0, max=0.99, step=0.01, continuous_update=False
),
noise=widgets.FloatSlider(
value=0.1, min=0, max=1, step=0.1, continuous_update=False
),
):
df = pd.DataFrame(columns=["feature1", "feature2", "target"])
# YOUR CODE HERE
raise NotImplementedError()
df["target"] = Y
df[["feature1", "feature2"]] = X
sns.scatterplot(data=df, x="feature1", y="feature2", hue="target", ax=ax)
plt.show()To normalize the features followed by DBSCAN, we create a pipeline as follows:
from sklearn.cluster import DBSCANdbscan_minmax_normalized = make_pipeline(
preprocessing.MinMaxScaler(), DBSCAN(eps=0.3, min_samples=3)
)
dbscan_minmax_normalizedTo generate the clustering solution, we can again use the fit_predict method as follows:
feature1, feature2 = df_spherical.columns[0:2]
cluster_labels = dbscan_minmax_normalized.fit_predict(
df_spherical[[feature1, feature2]]
)
plt.figure(num=3, figsize=(10, 5), clear=True)
_ = plt.subplot(121, title="Cluster assignment", xlabel=feature1, ylabel=feature2)
plt.scatter(df_spherical[feature1], df_spherical[feature2], c=cluster_labels)
plt.subplot(122, title="Cluster assignment", xlabel=feature1, sharey=_)
plt.scatter(df_spherical[feature1], df_spherical[feature2], c=df_spherical["target"])
plt.show()YOUR ANSWER HERE
@interact(
cluster_shape=["spherical", "non-spherical"],
eps=widgets.FloatSlider(
value=0.3, min=0.01, max=1, step=0.01, continuous_update=False
),
min_samples=widgets.IntSlider(value=3, min=1, max=10, continuous_update=False),
)
def cluster_regions_dbscan(cluster_shape, eps, min_samples):
df = {"spherical": df_spherical, "non-spherical": df_nonspherical}[cluster_shape]
feature1, feature2 = df.columns[0:2]
# YOUR CODE HERE
raise NotImplementedError()
plt.figure(num=4, figsize=(10, 5), clear=True)
_ = plt.subplot(121, title="Cluster assignment", xlabel=feature1, ylabel=feature2)
plt.scatter(df[feature1], df[feature2], c=cluster_labels)
plt.subplot(122, title="Cluster assignment", xlabel=feature1, sharey=_)
plt.scatter(df[feature1], df[feature2], c=df["target"])
plt.show()YOUR ANSWER HERE
OPTICS with Weka¶
For DBSCAN, the parameters and must be chosen properly. One needs to know how dense is dense enough to grow clusters, but this can be difficult, especially for high-dimensional data. A simpler alternative is to use OPTICS (Ordering points to identify the clustering structure):
We will use the package optics_dbScan in Weka for the density-based clustering algorithms. The package can be installed from Weka GUI -> tools -> Package manager.
Open the explorer interface and load the iris.arff dataset (not iris.2D.arff). Under the Cluster panel:
- Choose
OPTICSas theClusterer. - Choose
Use training setas theCluster mode. - Ignore the
classattribute using theIgnore attributesbutton. - Click
Start.
The OPTICS Visualizer will appear. The Table tab shows the list of data points in the order visited by the algorithm:
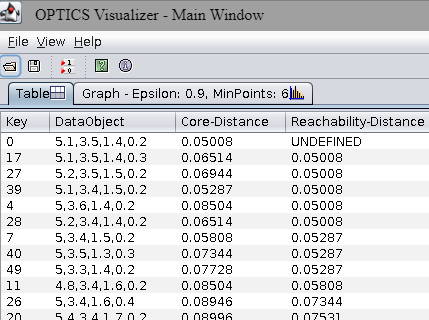
YOUR ANSWER HERE
The Graph tab shows the stem plots of core and reachability distances. We need to increase the Vertical adjustment in the General Settings panel to see the variations more clearly:
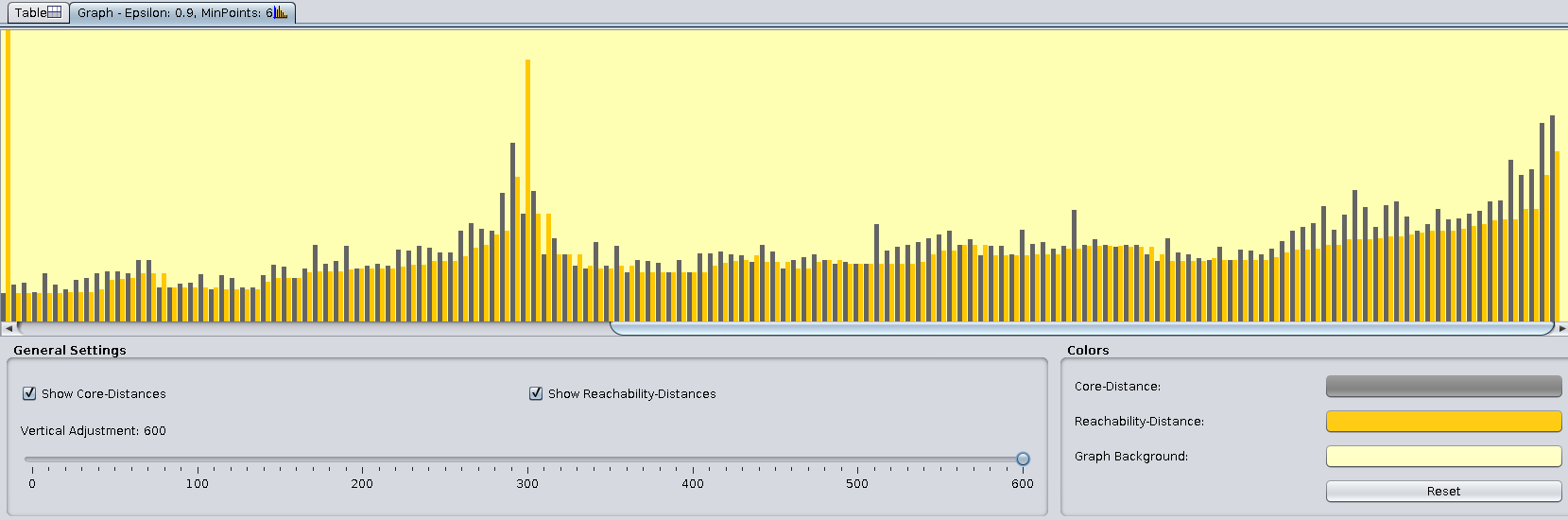
YOUR ANSWER HERE
Change the General Settings to give the reachability plot below:
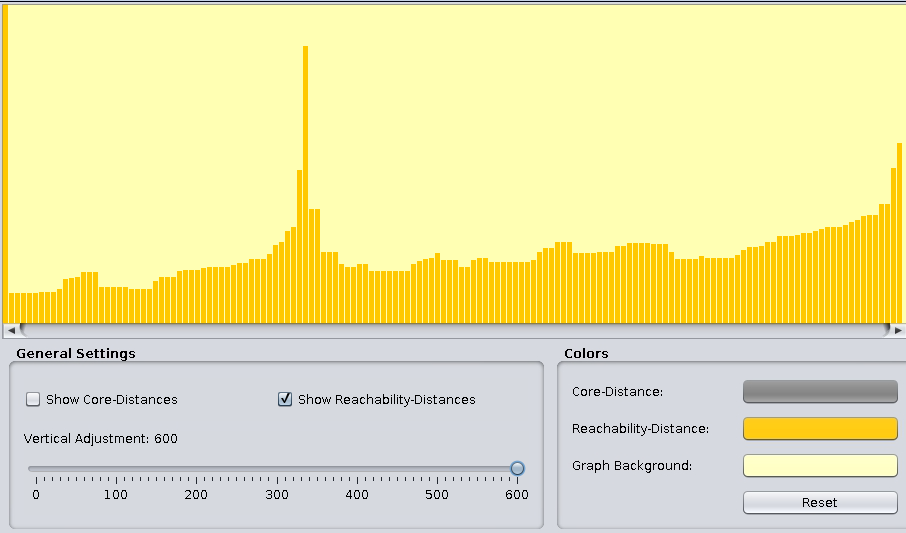
The above stem plot is called the reachability plot. To obtain a cluster from the plot,
- specify a threshold to clip the reachability distance from above, and
- identify a cluster as a “valley” of consecutively visited points with reachability distances below the threshold, except for
- the first point should have a reachability distance above the threshold.
- All other points not assigned to a cluster are regarded as noise.
# YOUR CODE HERE
raise NotImplementedError()
eps_clAssign to eps_cl a threshold value that results in 3 clusters. In particular, choose the threshold value that leads to as few noise points as possible.
# YOUR CODE HERE
raise NotImplementedError()
eps_clTo evaluate the 3 clusters obtained from a particular threshold using an extrinsic measure, run DBSCAN with
- the parameter
epsilonset to the threshold you obtained and - cluster mode set to
Classes to clusters evaluation.
Assign to error_rate the fraction of incorrectly classified instances and miss_rate as the fraction of instances not assigned to a cluster.
# YOUR CODE HERE
raise NotImplementedError()
error_rate, miss_rate